about
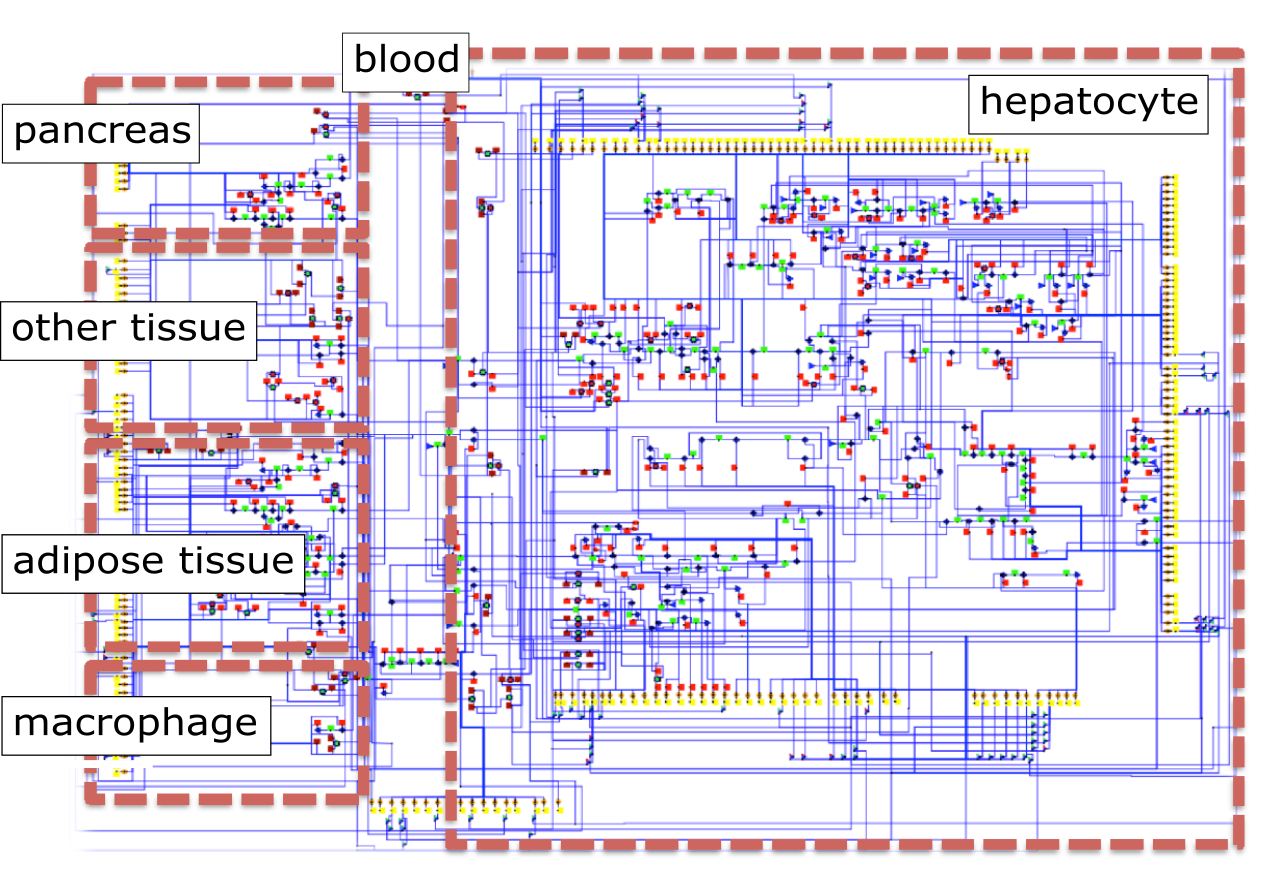
SysBio is a systems biology Modelica library that can be used to model large scale metabolic networks with object oriented approach. The library includes object classes corresponding to basic metabolic pathway entities, such as enzyme, metabolite, enzyme catalysed reaction, enzymatic mRNA, its transcription, etc. The derivation of these objects was performed for the normalised steady state, which is also the main limitation of the library. Due to this presumption, however, we can perform the simulations for metabolic networks even in the case of very sparse experimental data. The only kinetic data required to evaluate the normalised steady state of observed system are thus the distribution of fluxes in the metabolic network, the reversibily of enzyme catalysed reactions and the ratio between bound and free enzyme for each enzyme catalysed reaction. More information about the library and its establishment are available in the documentation page. An example of successful SysBio library application is the first integrated human metabolic model with multi-layered regulation, which was developed to investigate liver-associated pathologies, such as non-alcoholic fatty liver disease (NAFLD). The model is presented in the figure below and also available in the downloads page. You can find more details about the model in the following paper.